|
Gel-based oligonucleotide microarray approach to analyze ligand- and protein-DNA binding specificity
Laboratory of biological microchips also develops generic hexamer oligonucleotide platform which is primarily aimed at the study of sequence-specificity of ligands and proteins binding to ssDNA and dsDNA in vitro, particularly of proteins whose active sites bind relatively short stretches of DNA. Our 3D microarrays proved to be quite efficient for quantitative assessment of sequence specificity of low-molecular-weight compounds, ligands, and proteins in their interactions with ssDNA, see Figures 1 and 2. Such microchips may be used for the ranking of complete set of binding sequences in protein-DNA complexes.

|
| Click to enlarge.
|
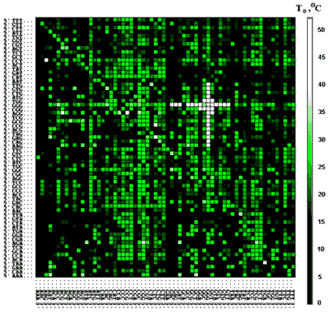
Figure 1. Computer representation of the data set for dissociation temperatures, TD, of the complexes between fluorescently labeled p50 protein (Y-box binding transcriptional factor) and octadeoxyribonucleotides immobilized on the generic microchip. Inner 6-mers of oligonucleotide 8-mers in this computer representation are arranged as two-dimensional matrix according to their 5'-halves in the rows and 3'-halves in the columns (Drobyshev et al., NAR, 1999). Color intensity of each small square reflects the TD value of the corresponding p50 - ss 8-mer complex. Color scale of the temperatures is shown on the right. P50 showed the highest sequence-specificity of binding to motifs 5'-NNGGGG-3', 5'-GGGGNN-3', 5'-NGGGGN-3', which are found in white cross on the matrice. (Zasedateleva et al., JMB, 2002).

|
| Click to enlarge.
|
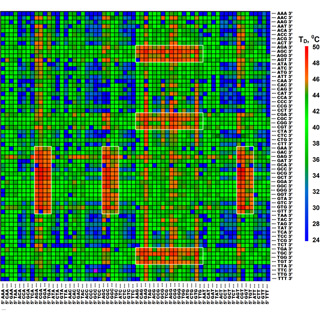
Figure 2. Computer representation of dissociation temperatures, TD, for complexes between TR-labeled RNase binase and octadeoxyribonucleotides of the generic microchip. Each hexanucleotide sequence should be read by combining its 5'-half in the column with 3'-half in the row. Color scale of TD values is shown next to the matrice. RNase binase showed the highest sequence-specificity of binding to motifs 5'-NG(A/T/C)GNN-3' and 5'-NNG(A/T/C)GN-3', which are found in six rectangular regions on the matrice. (Zasedateleva et al., NAR methods online, 2008).
In our platform single-stranded octamer oligonucleotides are immobilized within three-dimensional hemispherical hydrogel pads. The octanucleotides in individual pads 5´-{N}N1N2N3N4N5N6{N}-3´ consisted of a fixed hexamer motif N1N2N3N4N5N6 in the middle and variable parts {N} at the ends, where {N} represent A, C, G, and T in equal proportions. The chip has 4,096 pads with a complete set of hexamer sequences.
Immobilization of oligonucleotides in 3D-gel pads allows to register multiple temperature dissociation curves for protein-oligonucleotide complexes in parallel in equilibrium conditions and to define the affinity of a protein to ssDNA by comparing the obtained set of 46 = 4,096 dissociation temperatures: the higher the dissociation temperature of a given complex, the higher its binding affinity.
To study a protein in interaction with single stranded ssDNA the protein is labeled with a fluorescent dye. To study a protein in interaction with dsDNA an upper mixture of fluorescently-labeled complementary oligonucleotides (non-self-complementary) is hybridized with oligonucleotides of the microchip and dissociation of the duplexes is registered in presence and absence of the unlabeled protein. Affinity of a protein to dsDNA is assessed according to the shift of dissociation curve of a duplex, which occurs due to protein binding.
|


