Gel-based oligonucleotide microarray approach to analyze ligand- and protein-DNA binding specificity
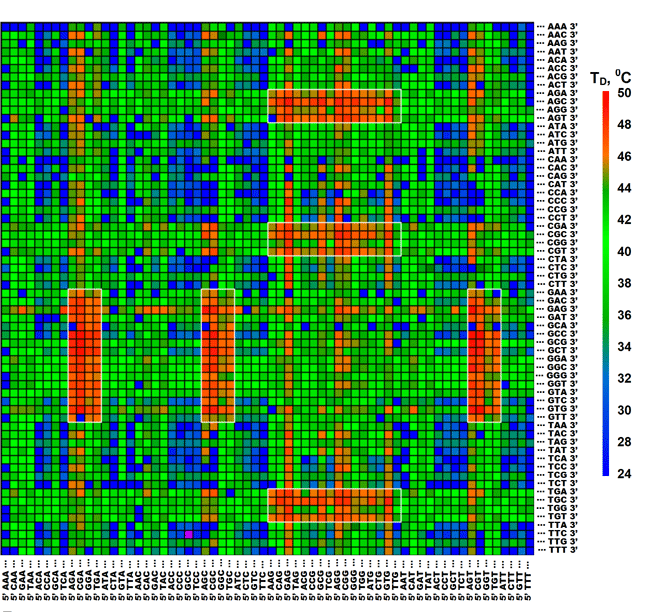
Figure 2. Computer representation of dissociation temperatures, TD, for complexes between TR-labeled RNase binase and octadeoxyribonucleotides of the generic microchip. Each hexanucleotide sequence should be read by combining its 5'-half in the column with 3'-half in the row. Color scale of TD values is shown next to the matrice. RNase binase showed the highest sequence-specificity of binding to motifs 5'-NG(A/T/C)GNN-3' and 5'-NNG(A/T/C)GN-3', which are found in six rectangular regions on the matrice. (Zasedateleva et al., NAR methods online, 2008).
|

